Aimer G. Diaz

Passionate about integrating computational and experimental approaches to unravel the complexity of plant-virus interactions and regulatory networks.
View My Scholar Google Profile
Research experience & Publications
Since the earliest steps of my education, I have been fascinated by basic research on virology and evolutionary biology, later in my career, I found out that the development and application of computational methodologies is of great help to answer questions on the intersection of these fields.
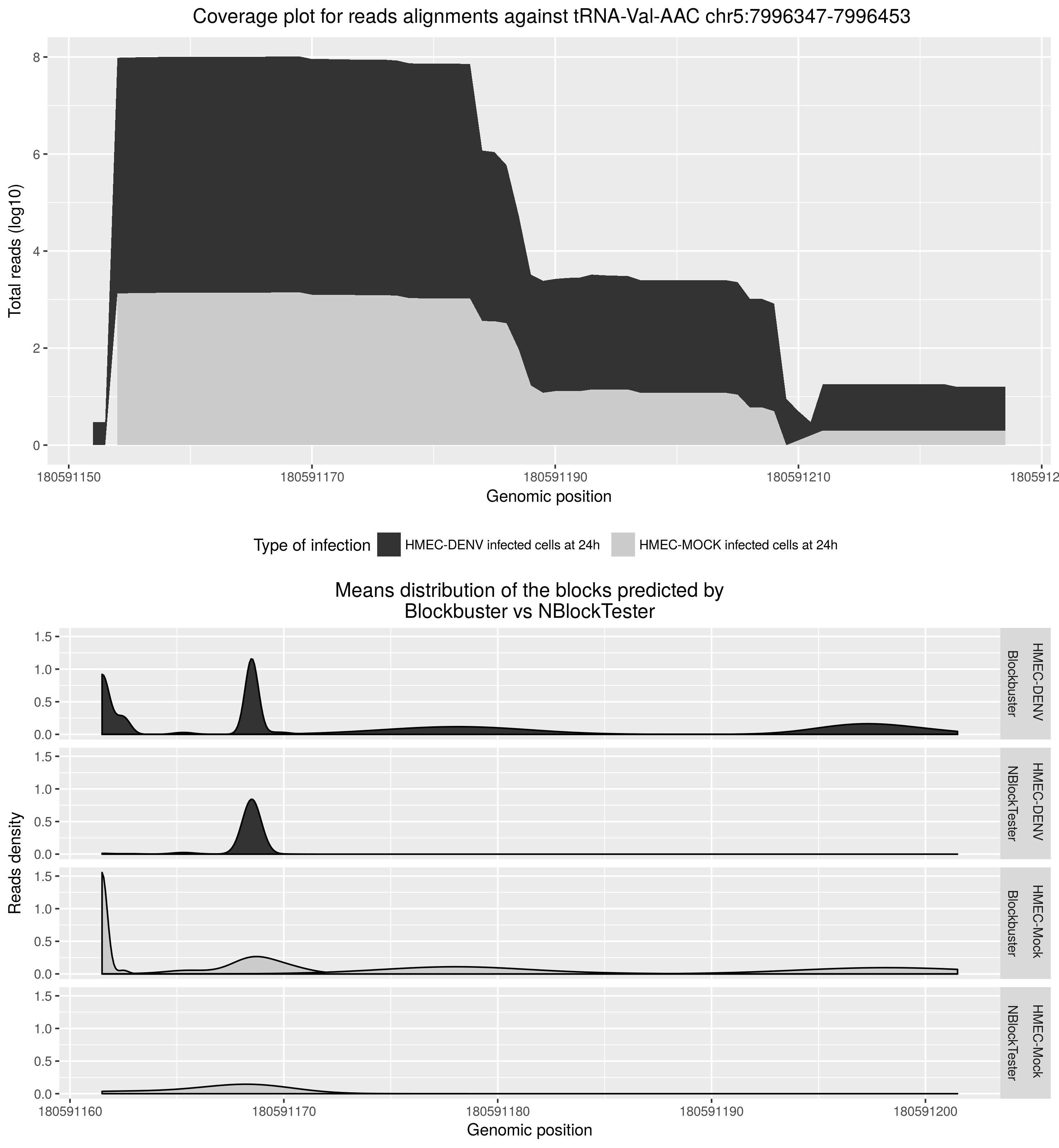
As a summary of my academic experience, during my master thesis in Dr. Clara Bermudez’s laboratory, I conducted computational analyses on small-RNAs dataset derived from multiple time-points of Dengue viral infected humans cell lines. In this project, I characterized Dengue-induce expression induction of miRNAs [1] and tRNA-derived fragments -a set of ncRNA molecules that required the development of new computational tools for their proper characterization. Specifically, I developed a bioinformatics tool called NBlockTester [2] to distinguish the expression of small RNAs from background noise in the ncRNA source.
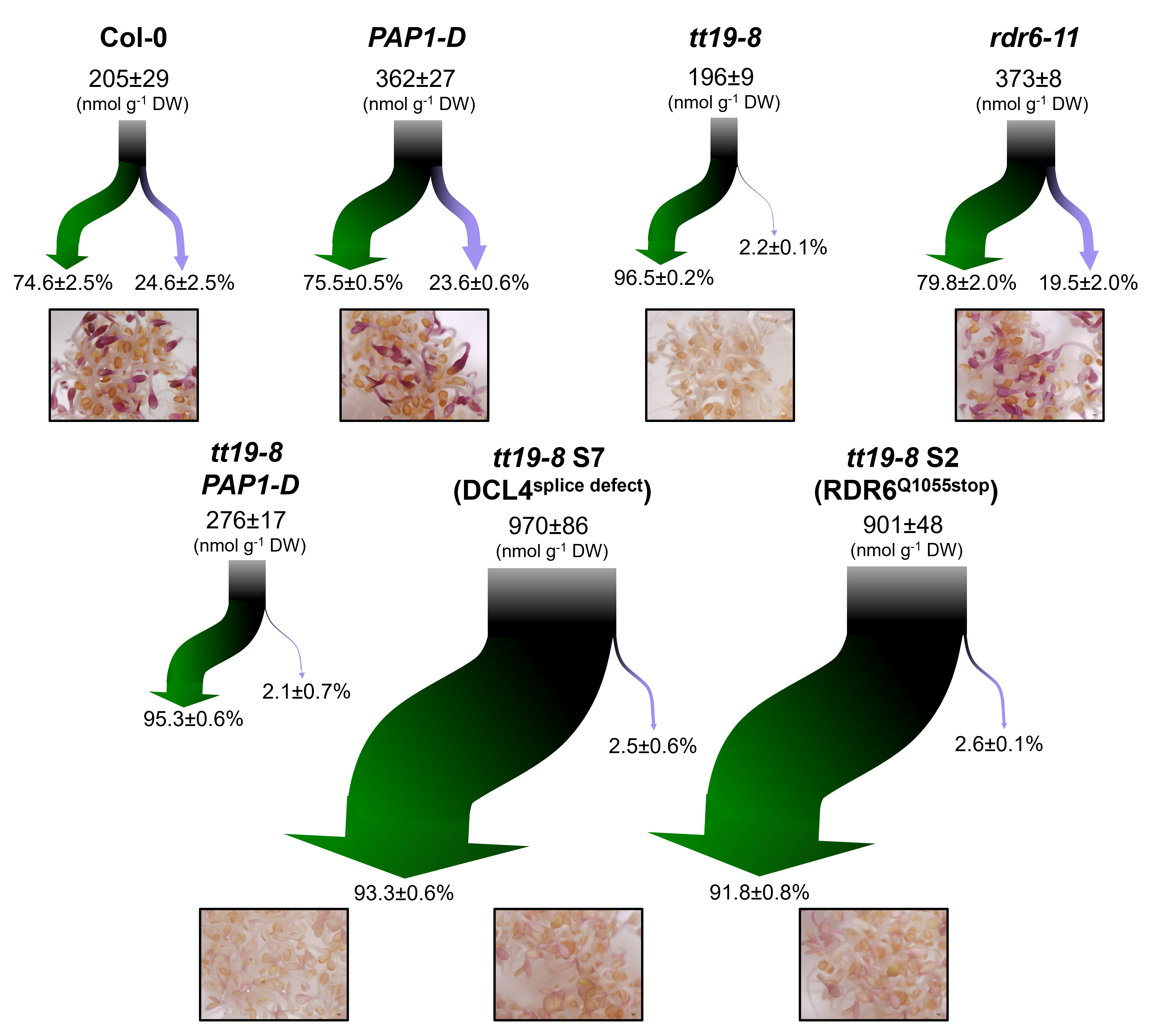
More recently, before beginning my Ph.D. in Sweden, I had the opportunity to work as visiting research in two laboratories. First in the Dr. Erich Grotewold lab, I worked with transcriptomic and small RNA-seq data to characterize a synergistic effect driven by the interfering role of trans-acting small RNAs in the regulation of specialized metabolism genes in Arabidopsis thaliana [3]. My second visiting research experience was in Dr. Adami’s lab, where I investigated the evolutionary persistence of a unique type of small ncRNAs found in Trypanosome brucei mitochondria, called guide RNAs. These molecules not only play a role in RNA editing of mRNAs but may also function as a mutational robustness system that protects the mitochondrial genome during severe and periodic bottlenecks.
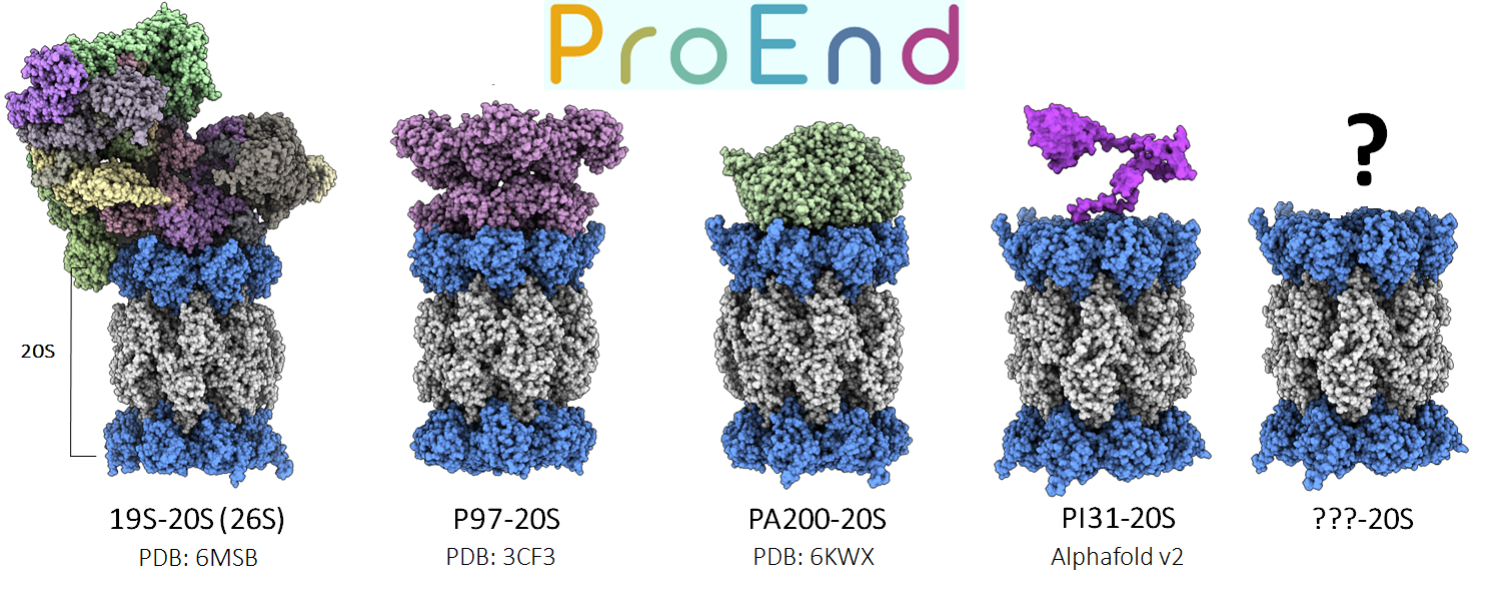
Lastly, In collaboration with David Salcedo at David Smith’s lab, I co-developed the ProEnd database, which analyzes HbYX motif-containing proteins across all life proteomes, of course including viruses [4]. Our work has focused on exploring the evolutionary and functional diversity of these motifs, particularly in relation to proteasome regulation. Through this collaboration, we have integrated bioinformatics approaches with experimental validation, identifying novel candidates that may expand the known roles of HbYX-like sequences.